3 BLAST: Introduction
The
BLAST(Basic Local Alignment Search Tool) software suite provides a set of tools for comparing a biological query sequence to the sequences in a database, and returning those sequences from the database that resemble the query sequence above a defined threshold similarity level.
The original BLAST paper is one of the most highly cited publications of all time, and has over 110,000 citations in the literature.
BLAST
- 1990:
BLASTis first described (Altschul et al. (1990)). - 1997: A refined version of
BLASTis published, introducing a new way of managing gapped alignments andPSI-BLAST- a method of compiling a profile of similar sequences to make searching more sensitive (Altschul et al. (1997)). - 2000: The
MegaBLASTalgorithm for fast alignment-based searching of large nucleotide sequences is proposed, and incorporated into theBLASTsuite (Zhang et al. (2000)). - 2009: A completely rewritten version of the software suite,
BLAST+is released. This improved performance and changed many features of the algorithms used for searching and building databases (Camacho et al. (2009)).
The latest updates to the BLAST software are described on the BLAST news page
3.1 Performing a BLAST search
This section describes a general BLAST query using the NCBI BLAST server. It is intended as a reference guide for you to return to as you get used to querying BLAST through this interface.
There are implementations of BLAST and other sequence search methods at many other databases, and they may present a different interface and choice of options, or even a totally different search method. For example, the RCSB-PDB protein structure database offers a sequence search page which uses the mmseqs2 search algorithm (Steinegger and Söding (2017)).
3.1.2 Select the BLAST tool you want to use
The BLAST suite provides search tools for finding matches to a query in a database. The query can be either a nucleotide or a protein sequence, and the database being searched can contain either protein sequences or nucleotide sequences. BLAST provides four different programs to carry out these combinations of search.
BLAST programs, and the combination of query/database sequence type they are used for
| Query type | nucleotideDB | proteinDB |
|---|---|---|
| nucleotide | blastn |
blastx |
| protein | tblastn |
blastp |
BLAST tools
The NCBI BLAST webserver provides specialised search options with specific combinations of parameters and databases pre-selected to support particular kinds of search (Figure 3.2).
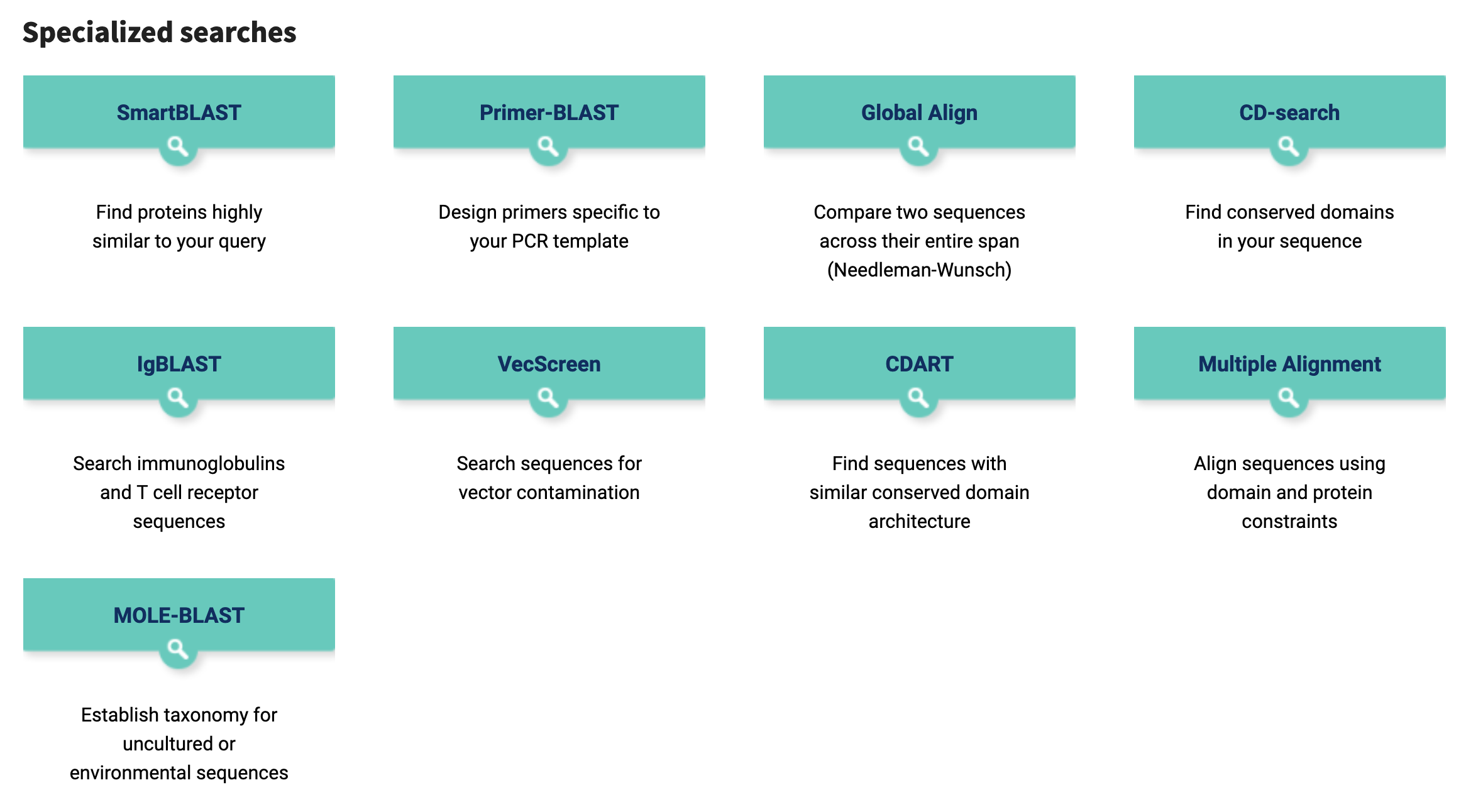
BLAST search options are available at the NCBI BLAST webserver
Select Nucleotide BLAST from the NCBI landing page, to get to the blastn search page (Figure 3.3).
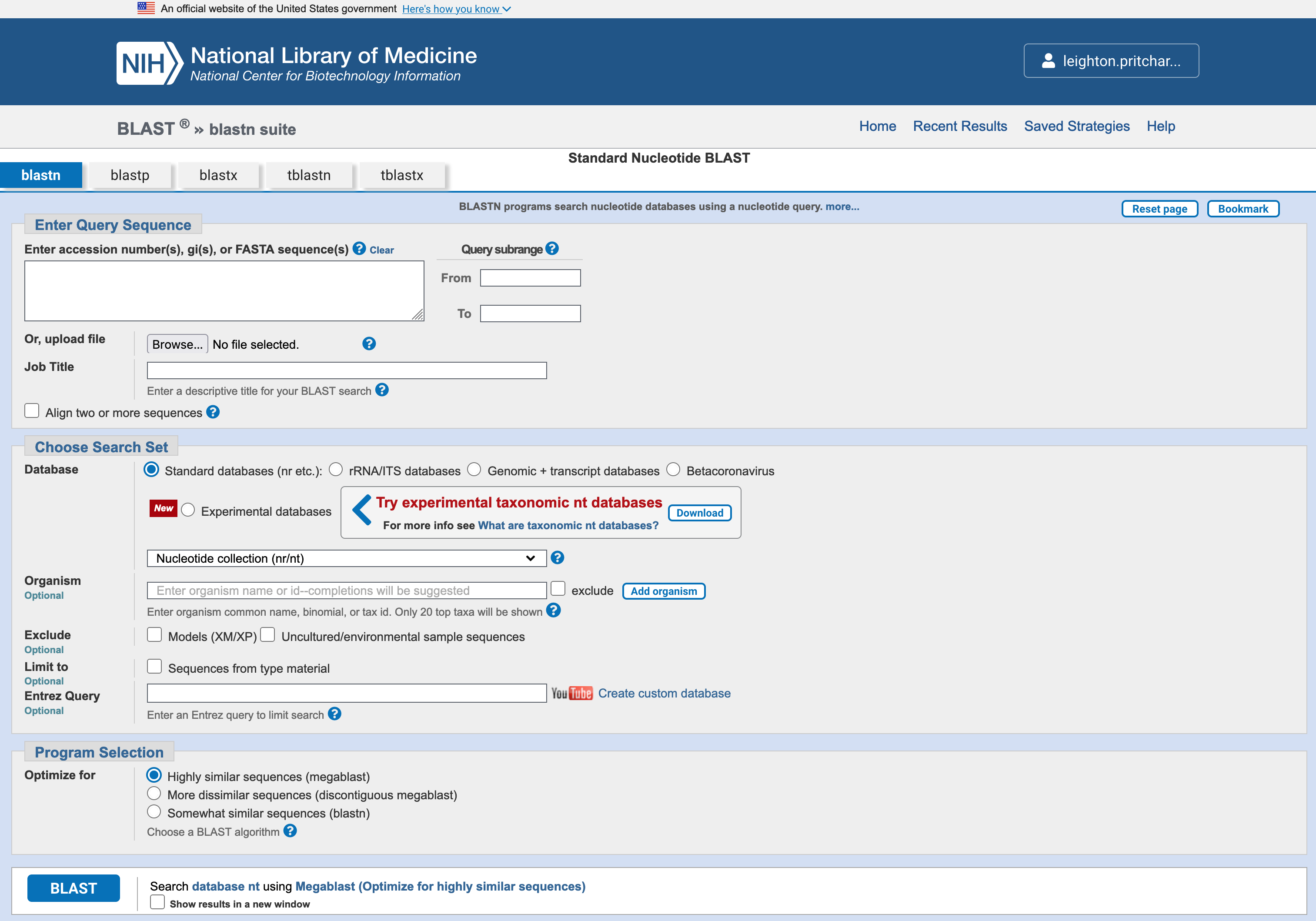
blastn webservice search page
3.1.3 Enter the query sequence
Copy the DNA sequence below, and paste it into the box marked Enter accession number(s), gi(s), or FASTA sequence(s) at the NCBI search page (Figure 3.4).
ATGCGTCGAGGGCGTCTGCTGGAGATCGCCCTGGGATTTACCGTGCT
TTTAGCGTCCTACACGAGCCATGGGGCGGACGCCAATTTGGAGGC
TGGGAACGTGAAGGAAACCAGAGCCAGTCGGGCC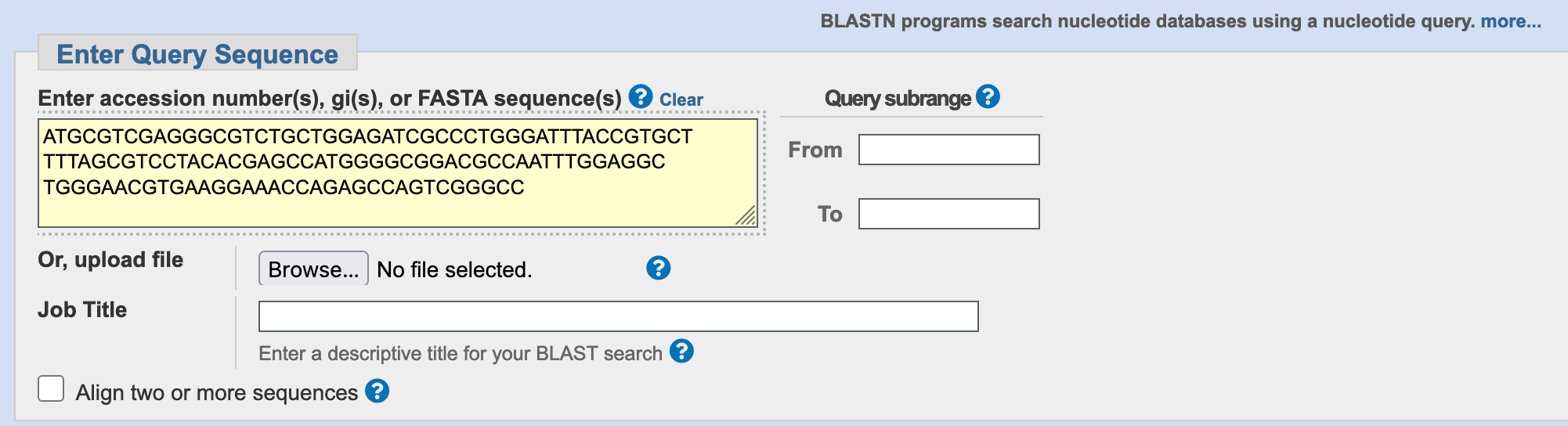
blastn search page with a query sequence pasted into the query sequence field.
3.1.4 Set appropriate parameter choices
If you make no more changes to the parameter settings for your search, the default options will be used. Your query will be made against the core_nt core nucleotide collection, a very large database. Due to the size of the database, the search may take a relatively long time.
You can make your BLAST searches quicker, and more relevant to your biological question, if you can use information about your sequence and the type of organism you want to search.
NCBI BLAST offers a number of smaller specialised databases with particular sequence types (e.g. RNA databases, sequences of protein structures, etc.) (Figure 3.5).
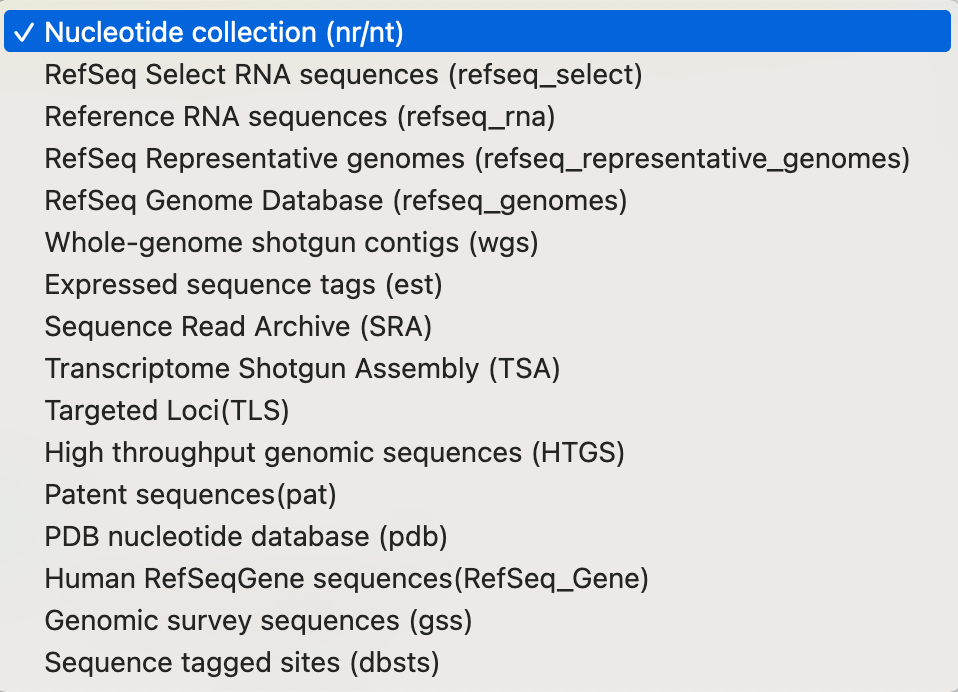
BLAST webserver
You can also narrow down the search by specifying an organism, or other taxonomic rank, using the Organism field (Figure 3.6).
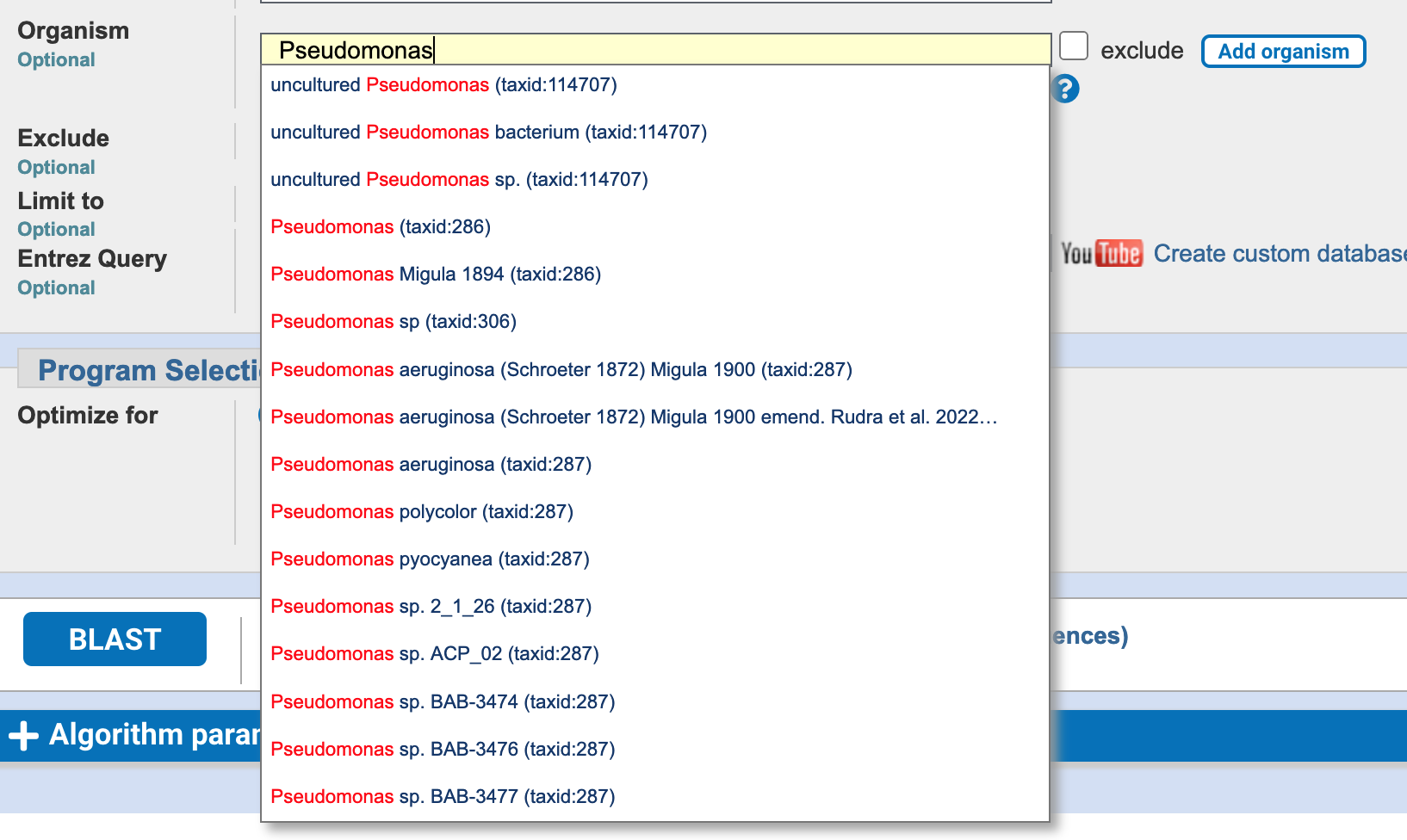
BLAST search organism field, for “Pseudomonas”
Restrict the sequences being searched by typing “Homo sapiens” in the Organism field and selecting the appropriate option from the drop-down list (Figure 3.7).
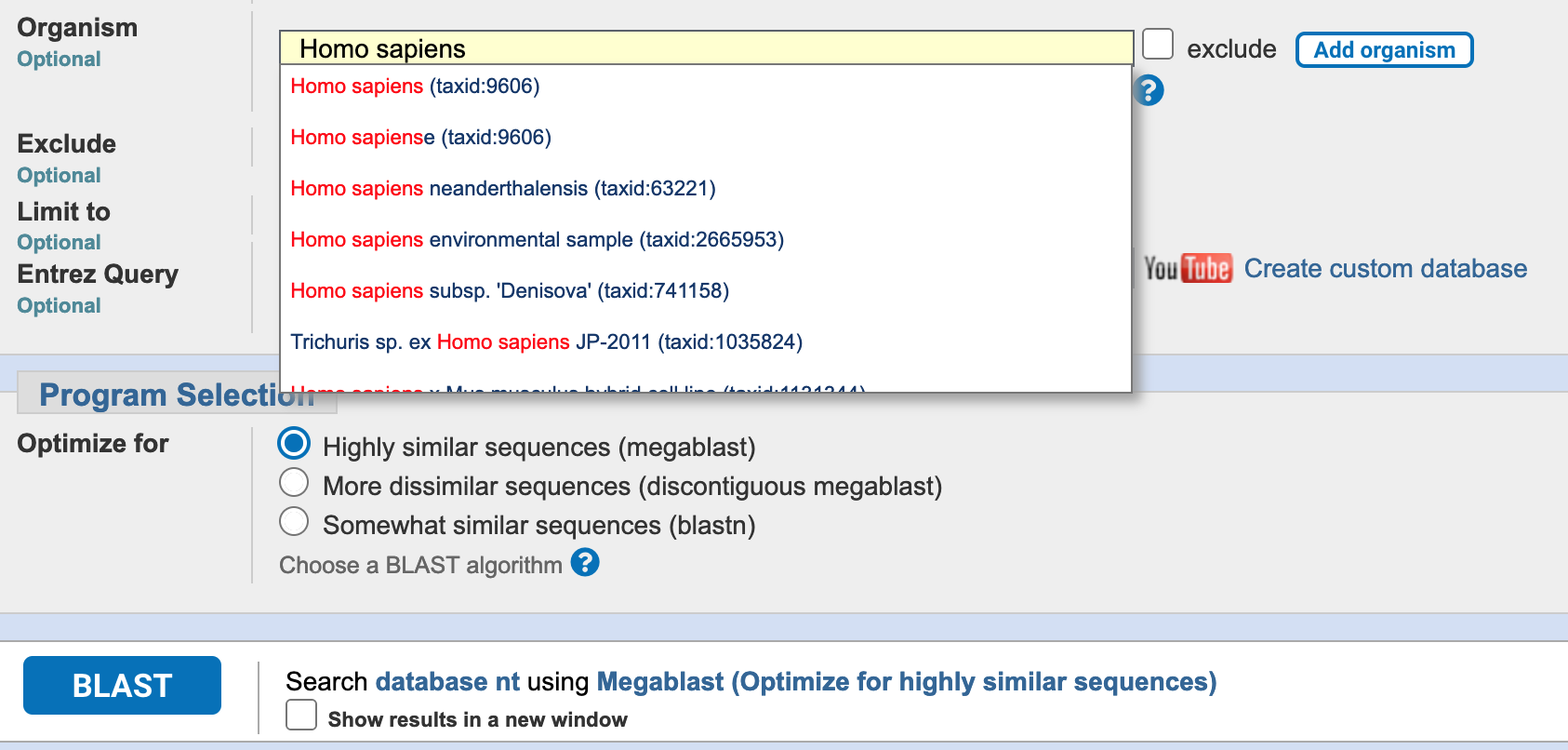
BLAST search organism field, for “Homo sapiens”
3.1.5 Run the BLAST search
Click on the BLAST button (Figure 3.8).
3.1.6 Wait for the search to complete
While the search runs, you will see a holding page that updates you with progress (Figure 3.9)
BLAST webserver progress page.
When the search is complete, you will see a blastn results page similar, but not identical, to Figure 3.10.
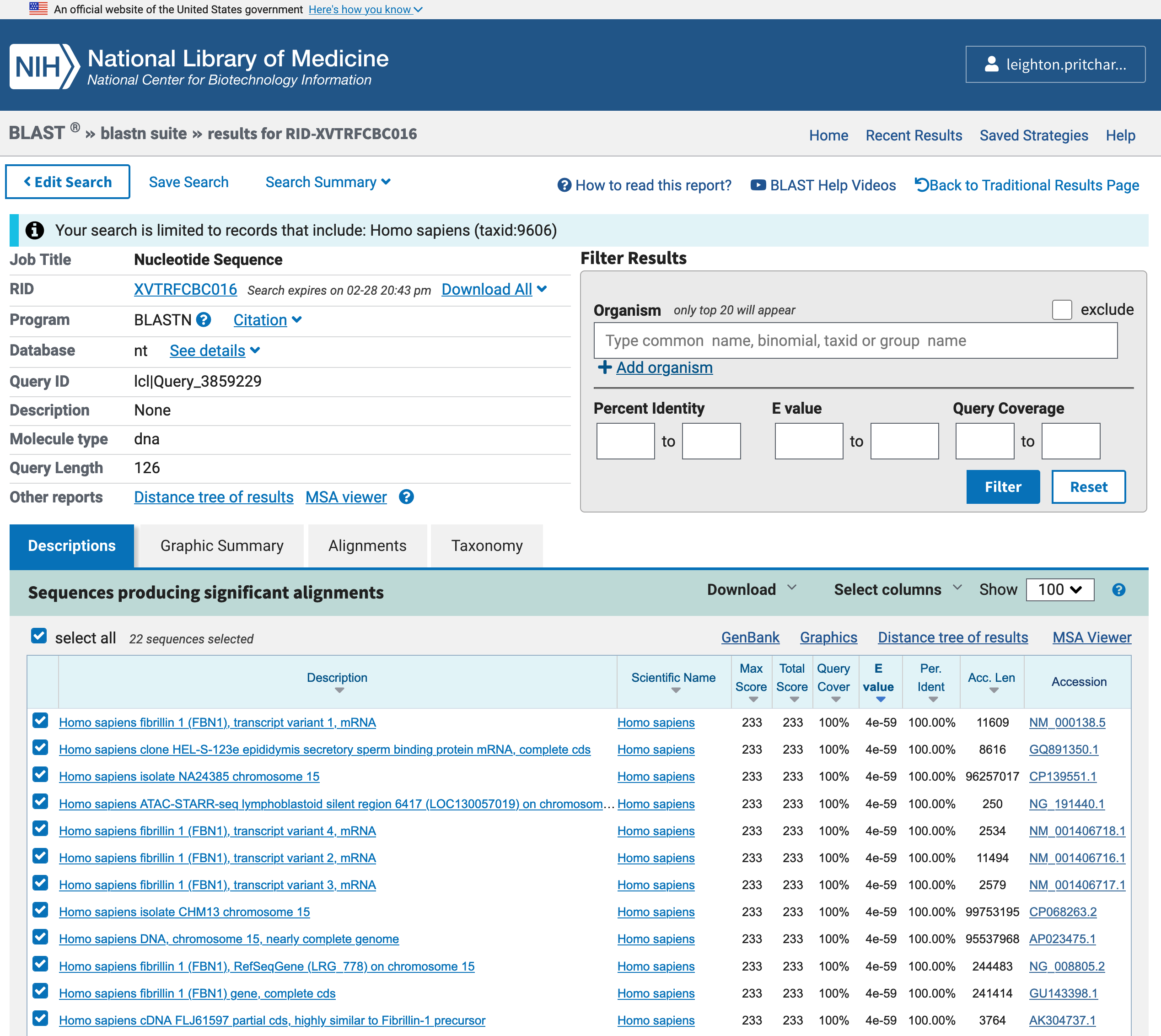
BLAST results page, for a blastn query.
3.2 Next Steps
After completing this section you should:
- be able to carry out a
BLASTsearch at the NCBIBLASTservice - be able to choose the correct database for your search type
- be able to modify search parameters, suitable for your biological question
Now you are ready to move on to using BLAST, which uses local sequence alignments to search large databases of reference sequences to find the best matches to an input query sequence.
You will next be presented with three scenarios, representing real uses of BLAST in research or clinical settings. Please work through the scenarios, completing the assessed sections and - if you have time - attempting the stretch activities.
There is a formative assessment on MyPlace for each scenario, which you should complete as part of the workshop.