5 Scenario 2: Protein Identification
5.1 Introduction
There has been an outbreak of diarrhoeal disease on a hospital ward, suspected to be due to a transmissible bacterial pathogen. You work in a clinical microbiology laboratory at the hospital, and you have been asked to help investigate.

Just as metagenomic analysis identifies which organisms are present in a sample, and gives an indication of their relative abundance, metaproteomic analysis does a similar job for the proteins that are present in a sample.
The human gut is a highly diverse microbial ecosystem populated by microbial communities that vary in composition along its length. Our understanding of these communities has been enhanced by metagenomic analyses that tell us which organisms are present in which communities, but also by metaproteomic analyses (e.g. Kolmeder et al. (2012)) which give insight into the molecular activities expressed by these communities.
Knowing what the normal, stable, gut microbial community looks like allows us to identify dysbioses - disturbances to the normal operation of these biological systems. MALDI-ToF (Matrix-Assisted Laser Desorption - Time of Flight mass spectrometry) is a valuable tool for identifying which organisms are present (e.g. Feucherolles et al. (2019)) and also for identifying and quantifying disturbances due to disease and measured as changes in the presence, absence, and abundances of proteins produced by the microbial communities (Debyser et al. (2019)).
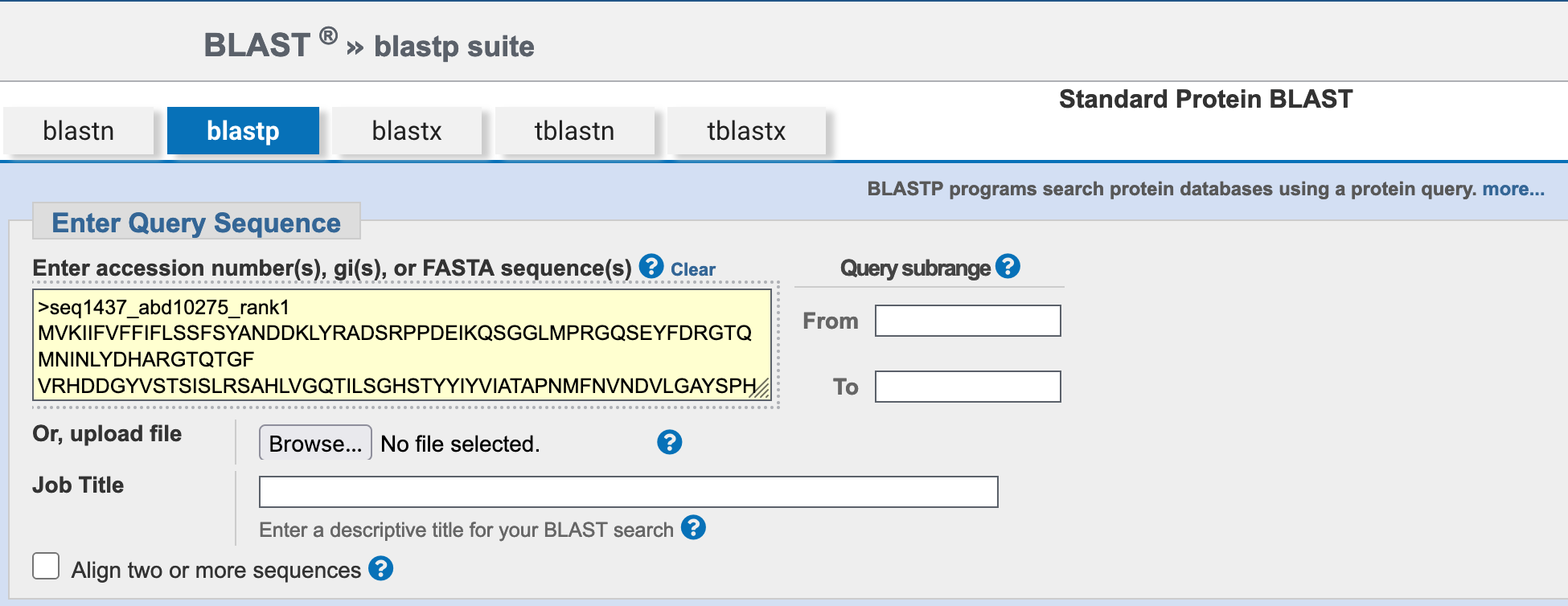
Using MALDI-ToF, you have carried out a metaproteomic analysis of one of the faecal samples, obtaining the protein sequence below as a highly-abundant protein, present in the sample.
>seq1437_abd10275_rank7
MVKIIFVFFIFLSSFSYANDDKLYRADSRPPDEIKQSGGLMPRGQSEYFDRGTQMNINLYDHARGTQTGF
VRHDDGYVSTSISLRSAHLVGQTILSGHSTYYIYVIATAPNMFNVNDVLGAYSPHPDEQEVSALGGIPYS
QIYGWYRVHFGVLDEQLHRNRGYRDRYYSNLDIAPAADGYGLAGFPPEHRAWREEPWIHHAPPGCGNAPR
SSMSNTCDEKTQSLGVKFLDEYQSKVKRQIFSGYQSDIDTHNRIKDELTo identify the potential function of this highly-abundant sequence, and the organism from which it comes, you want to BLAST the protein sequence against a suitable reference database at NCBI. Carry out this search using the guide below (using the hints if you need them) and answer the questions in the formative quiz on MyPlace.
5.2 Analysis Steps
Follow the steps below to carry out the analysis for this scenario. If you need a reminder, please check with the example search in the BLAST introduction chapter. If you need more help, the green boxes below expand to give you a hint for what to do, and the orange boxes expand to give a more direct instruction.
5.2.2 Select the BLAST Tool
BLAST tool should I use?
Your query sequence is a protein sequence, and you want to identify other protein sequences that have functional annotations, so this is a protein vs protein search.
For a reminder, see Table 3.1
BLAST tool to use
This is a protein vs protein search, so use the Protein BLAST tool.

5.2.3 Enter the query sequence
Paste the protein sequence above into the Enter Query Sequence field.
5.2.4 Set appropriate parameter choices
The NCBI BLAST webserver allows you to restrict your searches by taxonomic group.
The outbreak is thought to be caused by a transmissible bacterial pathogen, so perhaps you can restrict your search to a subset of a larger, comprehensive database?
The NCBI BLAST webserver allows you to restrict your searches by taxonomic group. You can query within the database, while restricting your search only to proteins that come from bacteria.
Select the nr_cluster_seq database, and select Bacteria from the Organism field
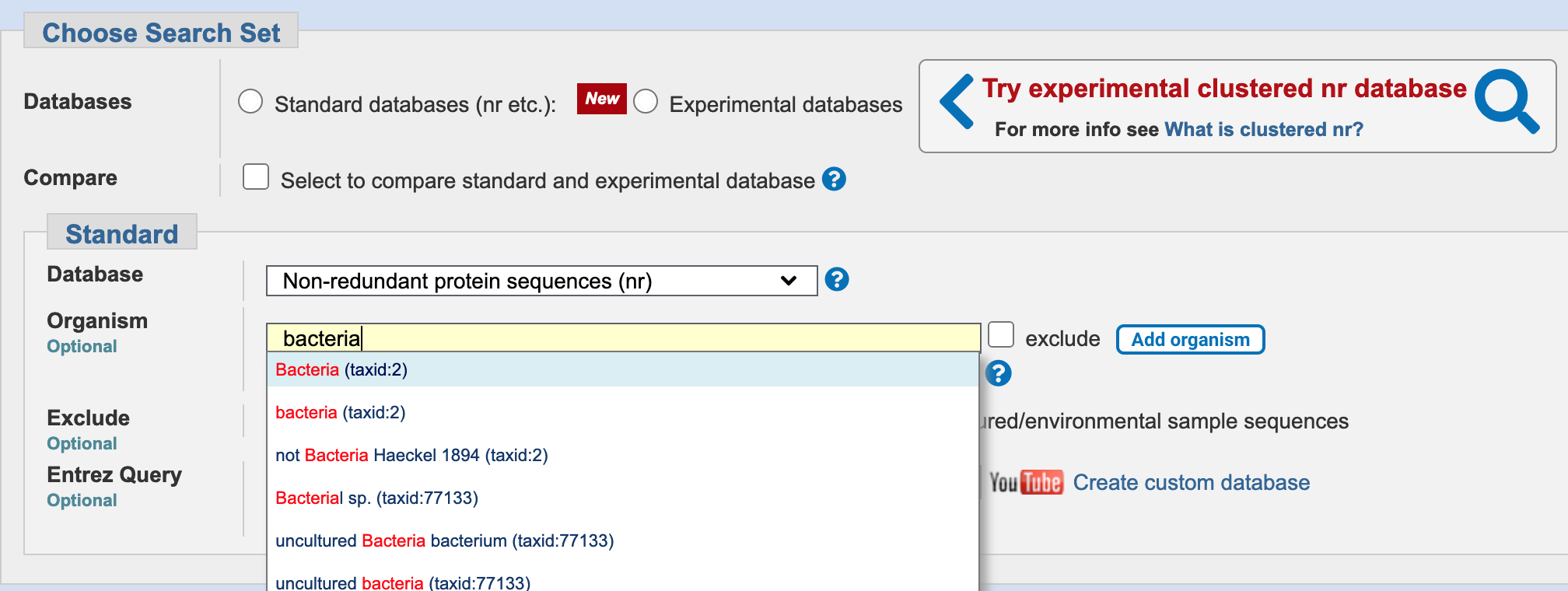
nr database, filtering on bacteria
5.2.5 Run the BLAST search
BLAST search
Click on the BLAST button.
5.2.6 Interpret the BLAST report (MyPlace Questions)
It is not unusual to find that a query matches many proteins in the database. By identifying those most closely-related to the query sequence, you should be able to establish a likely function for your protein, and a candidate identity for the organism it originates from.
Please answer the questions below in the formative quiz on MyPlace
Clicking on the green box should give you a hint to the answer, or where to find it.
Check the Accession column under “Sequences producing significant alignments” in the report’s Descriptions tab.
Check the Description column under “Sequences producing significant alignments” in the report’s Descriptions tab.
Check the Total Score column under “Sequences producing significant alignments” in the report’s Descriptions tab.
Check the Query Cover column under “Sequences producing significant alignments” in the report’s Descriptions tab.
Check the Per. Ident column under “Sequences producing significant alignments” in the report’s Descriptions tab.
Click on the accession number for the best match. This will open a new tab in your browser with the gene record.
The gene record contains an annotation field called /product that can be found just above the sequence data. If you cannot see it, search for /product in the page.
BLAST report a hit?
Scroll down the table in the Descriptions tab, or use the Taxonomy tab to find other organisms
Use the Graphic Summary tab to visualise the pairwise alignments, and examine the superfamily information in the graphic at the top of the page.
5.3 Stretch Activities
These activities are not necessary for the assessment, but provide more information relevant to interpreting your BLAST results. Open the orange boxes to see the questions, and use the green boxes to find some answers.
5.3.1 Find out more about your protein’s function
Graphic Summary tab in the report
BLAST reports often link to external sources of data and information that can help you interpret your results. At the top of the Graphic Summary tab there is a clickable graphical indicator of a putative conserved domain in your protein. Click on the green box. This will open an entry in the NCBI Conserved Domain Database (CDD).
Click on the small [+] symbol at the left of the entry to see an alignment of your protein against the canonical sequence for the conserved domain.
Conserved domains are usually associated with Protein Family (PFam) domain records. This is also true for the matches to your protein. Click on the link to the PFam entry (in the Accession column of the CDD record) to be taken to a multiple sequence alignment of representative sequences for this domain, which opens in a new tab.
To see the PFam record itself, click on the Source: pfam link in the Links box at the top of this page. This opens the InterPro PFam page with more information about the function and structure of the conserved domain in your protein.
5.3.2 Inspect the match taxonomy
Distance tree of results link in the report
The Distance tree of results link will open a new tab containing a phylogenetic tree, showing the placement of your sequence in relation to the matching sequences. Some questions to consider:
- Are the results in the tree consistent with the results in the
Descriptionstab? Is there any extra information here that is not found in theDescriptionstab?
Distance tree
- Yes, they are consistent. The tree gives us evolutionary information about the order of divergence of each organism’s sequences, when they diverged, and also which organisms’ sequences are more similar to each other.
5.3.3 Repeat the search with different parameters
Return to your BLAST query by clicking on the Edit Search button at the top of the report.
BLAST report Edit Search button
Use the nr database rather than the default choice. Check the Exclude box next to the Organism field, to exclude bacterial sequences from your BLAST search.
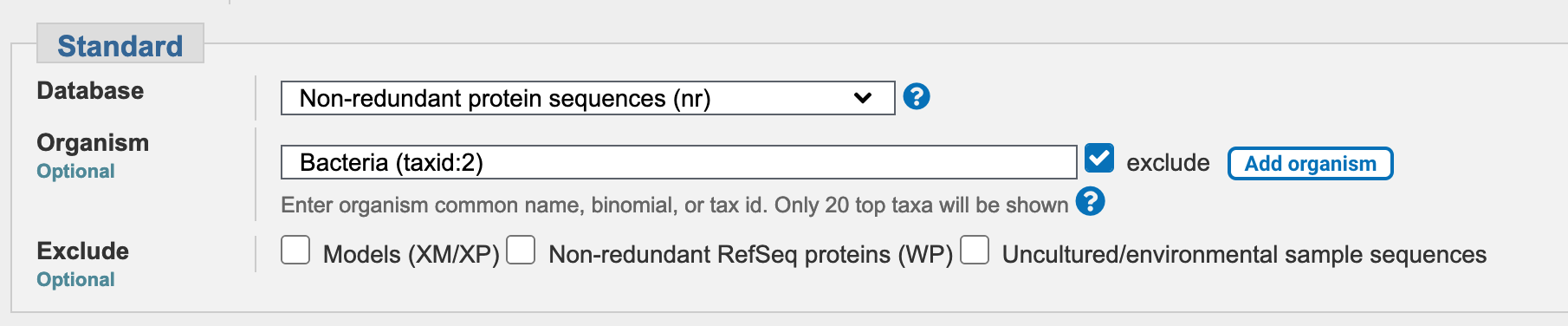
BLAST search
Click on the BLAST button.
- Are there any exact matches to your query sequence in the
BLASTresults? - If there are exact matches to your query, what kind of organism are they found in?
- Why might this protein sequence be associated with that kind of organism?
- Yes, there is one exact match to a non-bacterial sequence.
- The sequence is found in a virus (and very similar sequences are also found in other viruses)
- Pathogen virulence genes are often transferred between bacteria by viruses or phage infection in a process called horizontal gene transfer (HGT).
5.4 Summary
After successfully working through this scenario, you should be able to
- identify the putative function of a protein and the organism from which it is likely to originate
- identify and name conserved domains of a protein, using the
BLASToutput
If you completed the stretch activities you should be able to
- use the graphic summary to identify more information about the conserved domains, using the CDD database
- find and explain the
BLASTresult’s phylogenetic tree and taxonomy information, and use it to understand the relationships between the matching sequences - modify your search parameters for the same query, using taxon filters to exclude organisms from your search and identify biologically-useful information