1 16S Sequence Taxonomic Identification
1.1 Introduction
In this exercise you will use the 16S sequence that you amplified from your unknown bacterium in order to make a taxonomic identification.
To do this, you will use two online bioinformatics services:
- NCBI’s
BLASTtool (you have already met this in Workshop 2) silva, a quality-checked and curated 16S sequence database service

It might tempting to take computational results at face value, and to believe that the “computer can’t be wrong.” But, in bioinformatics as elsewhere in life, computers and databases can sometimes be in error or give imprecise answers. In general the results of a sequence database search, like querying a 16S sequence against a public database, depend on a number of factors, including:
- the query sequence (obviously)
- the contents of the database
- the search tool used to query the database
- the parameter settings for the search tool
and even if the query sequence is the same, using a different service will often change at least one of the other three factors - and maybe other thigs besides.
This exercise will give you experience in using alternative bioinformatics tools for the same task.
1.2 Your data
You should download the 16S sequence file you obtained from your experiment in Laboratory 3.
This will be a plain-text file in FASTA format.
1.3 Analysis 1: NCBI BLAST
Follow the steps below to carry out a blastn search using your 16S sequence data.
1.3.2 Select the BLAST Tool
BLAST tool should I use?
Your query sequence is a nucleotide sequence, and the 16S sequences of other organisms are also nucleotide sequences, so this is a nucleotide vs nucleotide search.
For a reminder, see Workshop 2.
BLAST tool to use
This is a nucleotide vs nucleotide search, so use the Nucleotide BLAST tool.

1.3.3 Enter the query sequence
You can enter the query sequence in either of two ways.
- Open your 16S sequence file in a text editor, copy the data, and paste the data into the
Enter Query Sequencefield.
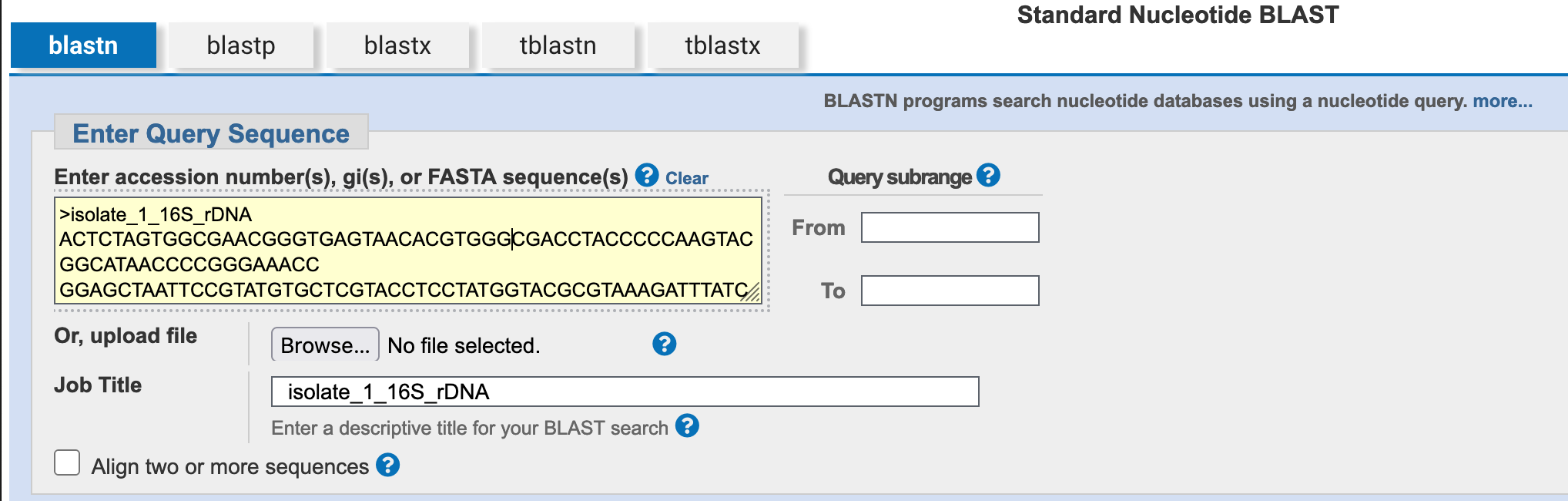
- Click on the
Browse…button to open a file selection dialogue box, then select your 16S sequence file. The uploaded file’s name will then be visible next to theBrowse…button.
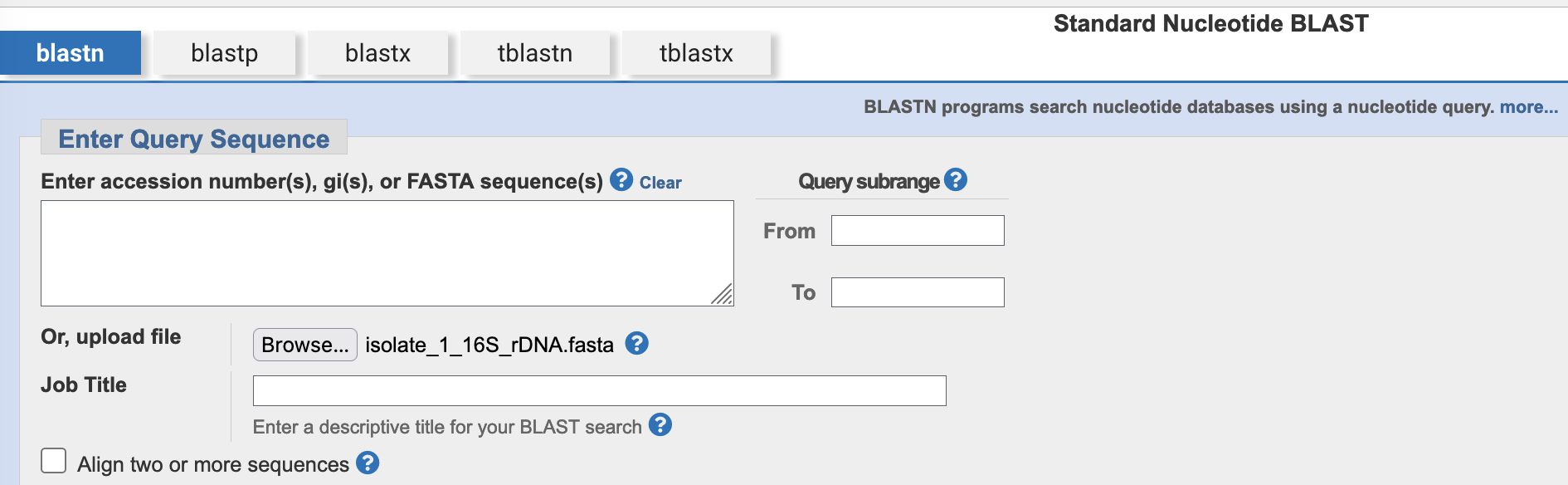
Browse… button
1.3.4 Set appropriate parameter choices
The NCBI BLAST webservice provides specialised databases for common, well-defined search tasks.
You have a well-defined set of target sequences that you want to compare your query with: they are all 16S sequences. Can you see a database on the search page that might be suitable?
The NCBI BLAST webservice provides specialised databases for common, well-defined search tasks, including searching for 16S or ITS (Internally-Transcribed Spacer) sequences when identifying unknown microorganisms.
Select the rRNA/ITS databases radio button, and ensure that the “Bacteria and Archaea” database is selected.

1.3.5 Run the BLAST search
BLAST search
Click on the BLAST button.
1.3.6 Interpret the BLAST report (MyPlace Questions)
It is common for 16S rDNA sequences to match many other bacterial sequences in the database. However, by identifying those most closely-related to the query sequence, you should be able to establish a likely identity for this organism.
As you all have different sequences there is no MyPlace quiz, please answer these questions for yourself.
If you would like guidance about your answer, please ask one of the tutors in the workshop.
Clicking on the green box should give you a hint to the answer, or where to find it.
Check the Accession column under “Sequences producing significant alignments” in the report’s Descriptions tab.
Check the Description column under “Sequences producing significant alignments” in the report’s Descriptions tab.
Check the Total Score column under “Sequences producing significant alignments” in the report’s Descriptions tab.
Check the Query Cover column under “Sequences producing significant alignments” in the report’s Descriptions tab.
Check the Per. Ident column under “Sequences producing significant alignments” in the report’s Descriptions tab.
1.4 Analysis 2: silva
silva is a curated, quality-checked database of rRNA sequence data that has been run by the Liebniz Institute DSMZ German Collection of Microorganisms and Cell Cultures for about two decades. The silva site provides a number of online tools and services, including the ACT service that enables users to search the database with their own rRNA sequences to find the best matches and identify their organism.
silvarRNA database projectsilvaAlignment, Classification, and Tree (ACT) webservice- ACT online tutorial
1.4.2 Enter the query sequence
You can enter the query sequence in either of two ways.
- Open your 16S sequence file in a text editor, copy the data, and paste the data into the
Input datafield (where it says “Paste your FASTA sequence here”).
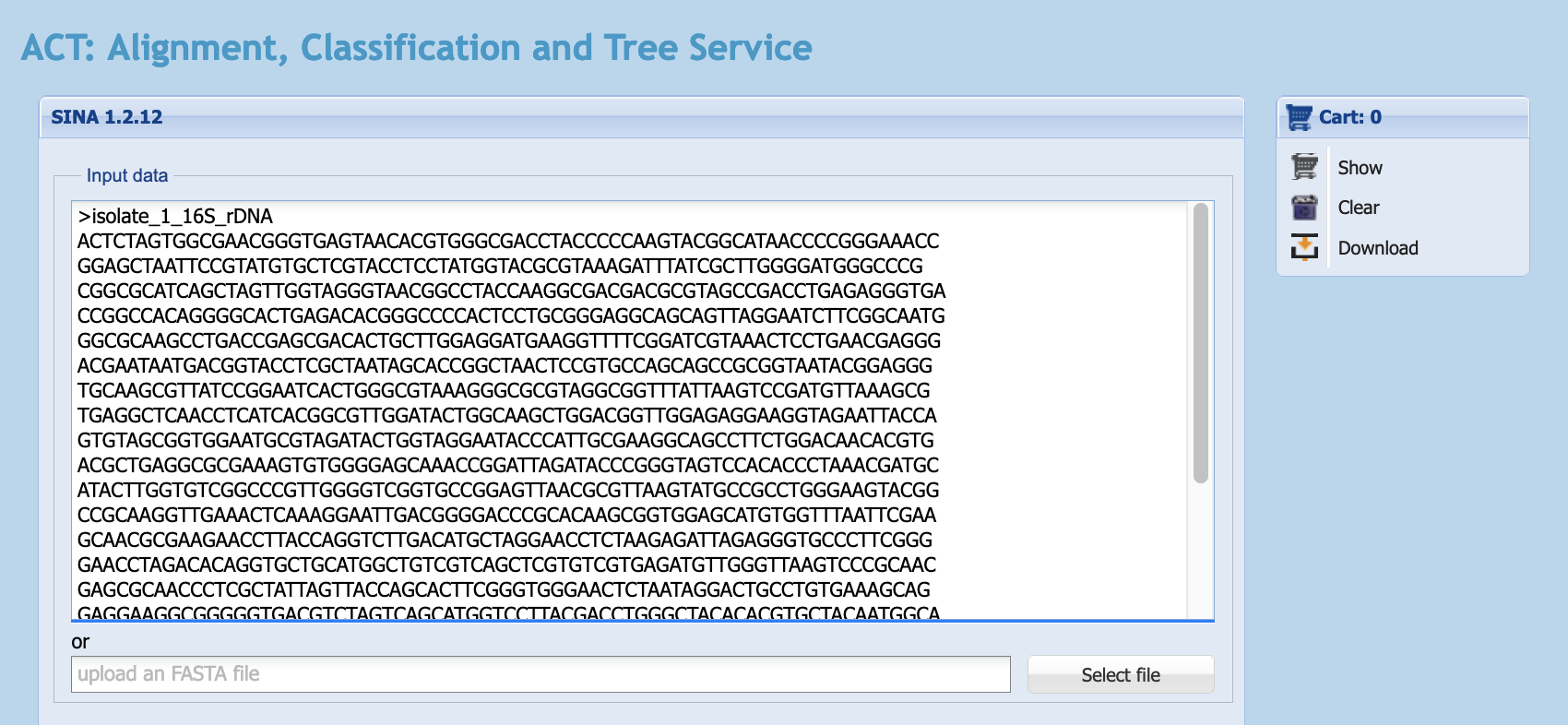
silva landing page with sequence pasted into the Input data field
- Click on the
Select filebutton to open a file selection dialogue box, then select your 16S sequence file. The uploaded file’s name will then be visible in the field next to theSelect filebutton.
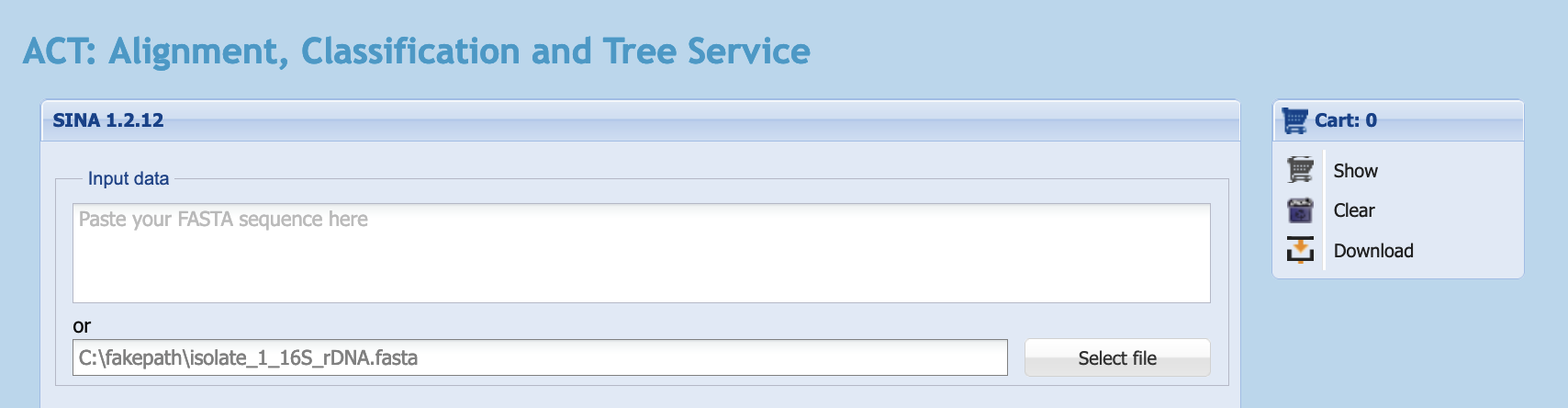
silva landing page with sequence uploaded as a file
1.4.3 Set appropriate parameter choices
silva is designed specifically to assign taxonomy from 16S sequence data, so you shouldn’t usually need to modify any parameters. However, you do need to tell silva what actions you want it to take.
You want to search with your sequence and classify it.
You need to tell silva that you want to search with your sequence and classify it. So check the box marked Search and classify.
The default settings (95% minimum sequence identity, reporting ten neighbours) are a good starting point.
Once you select Search and classify, a new set of options will appear in the window (Figure 1.10) and these can also be left as they are.
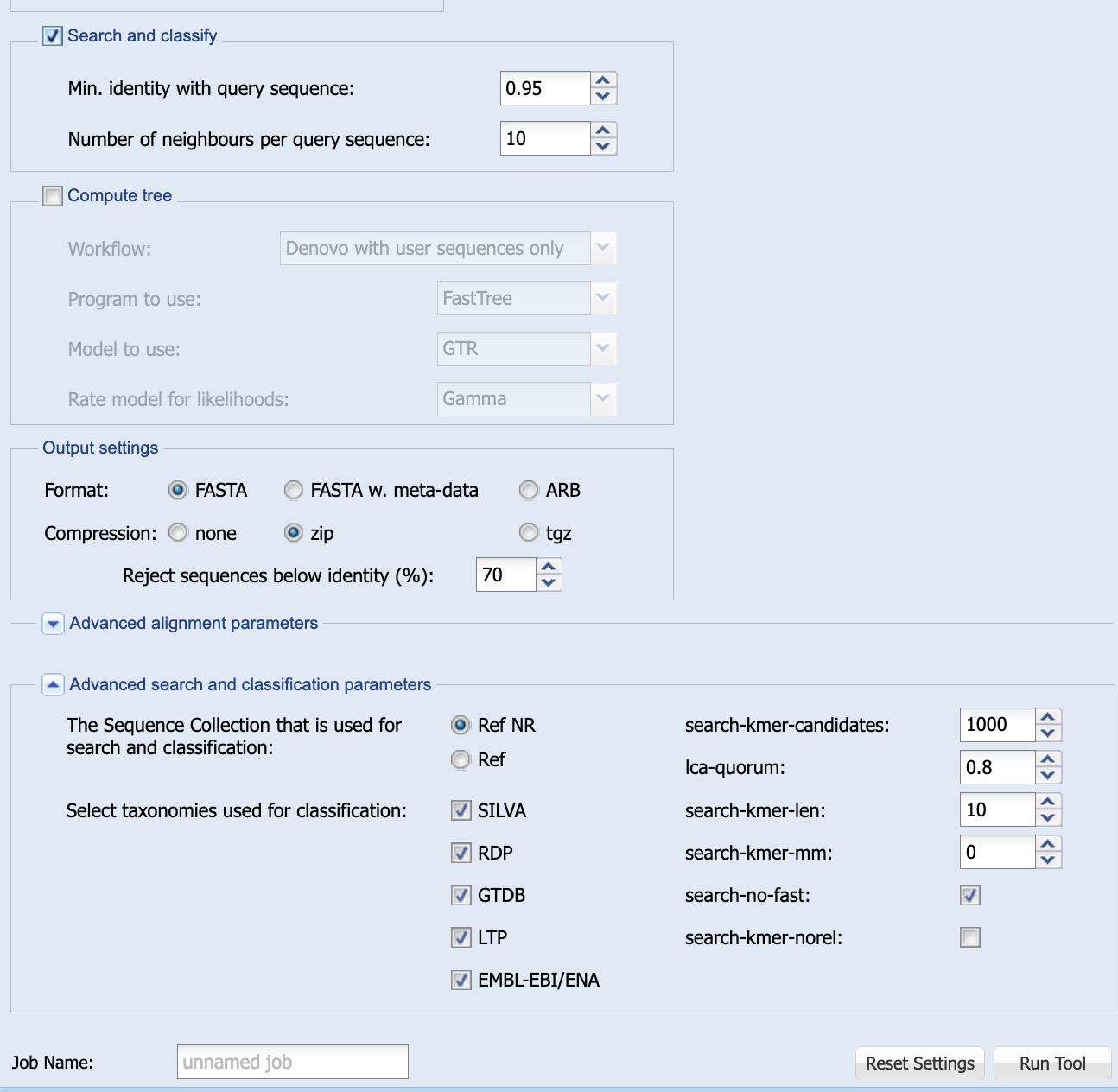
silva’s default Search and classify options.
1.4.4 Run the silva search
silva search
Click on the Run Tool button (lower right of Figure 1.10).
silva does not provide results as quickly as NCBI’s BLAST server. Keep your eye on the Aligner Taskmanager section of the page, to monitor progress.
silva’s progress will run through several stages, from initialising to finished.
silva’s Aligner Taskmanager table
1.4.5 Interpret the silva report (MyPlace Questions)
Unlike BLAST, silva provides the identity of the last common ancestor (LCA), rather than reporting a sequence search match. It displays results in the Alignment Result Table (Figure 1.12). This table displays scores but, by clicking on the Display Classification button, you can see the taxonomic identification silva has made for your sequence (Figure 1.13).
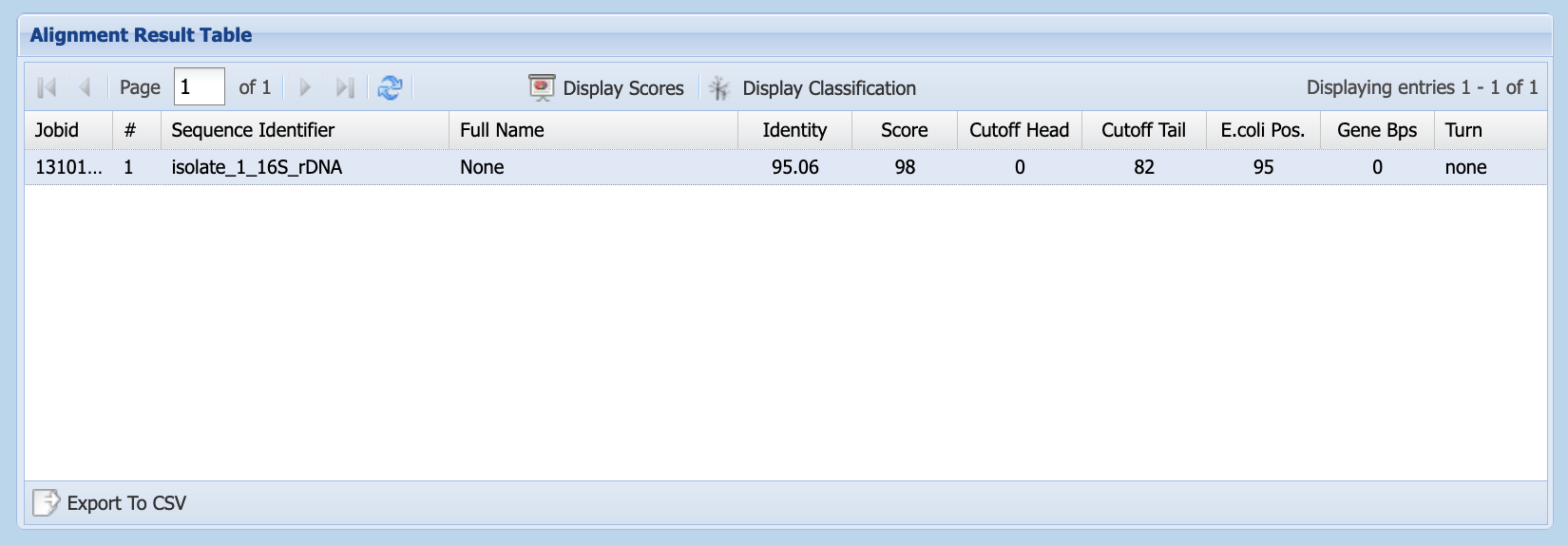
silva’s Alignment Result Table
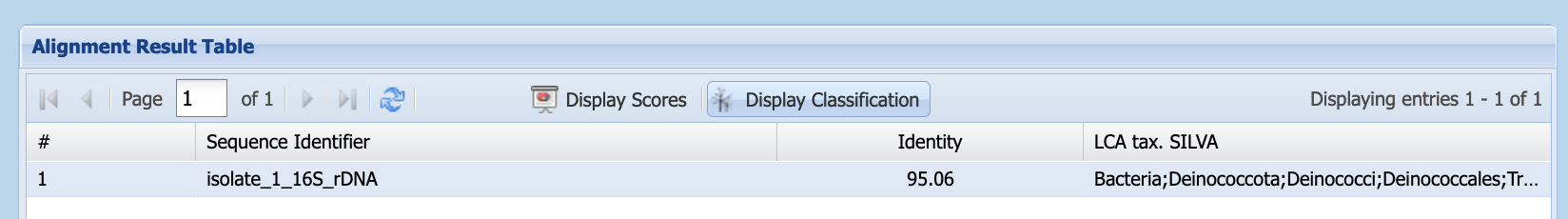
silva’s Alignment Result Table, showing classification information
Use the “handles” to drag the size of the column headers to the left or right, until you can see the classification, as in Figure 1.14.
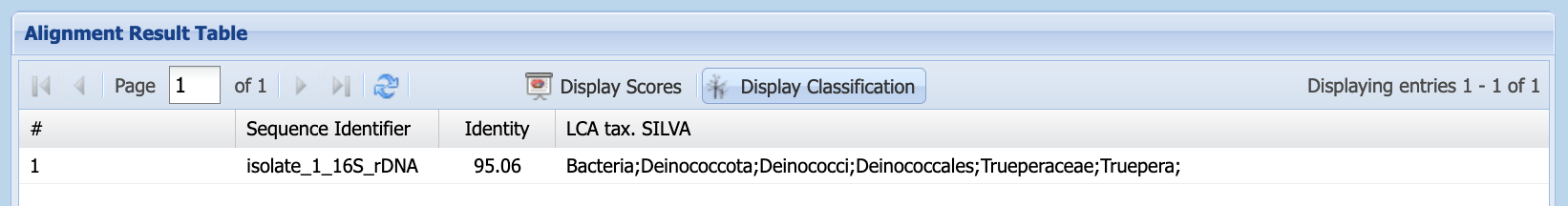
silva’s Alignment Result Table, showing classification information with wider columns
Alternatively, use the Export to CSV option to download a plain-text .csv file containing silva’s results for your sequence. You can open this in a text editor, or in Excel.
As you all have different sequences there is no MyPlace quiz, please answer these questions for yourself.
If you would like guidance about your answer, please ask one of the tutors in the workshop.
Clicking on the green box should give you a hint to the answer, or where to find it.
silva assigns to your organism?
You can find this in the LCA tax. SILVA column of the Alignment Result Table, or in the lca_tax_slv column of the downloaded .csv file.
You can find this in the Identity column of the Alignment Result Table, or in the identity column of the downloaded .csv file.
BLAST and silva searches give the same taxonomic identity for your 16S sequence?
Inspect the outputs of both methods, and compare the taxonomic assignments.
Among other things, you may want to consider:
- did both tools give the same identification?
- how much sequence identity was shared between your query sequence and the matches each tool found?
- is your organism common enough that there are many examples of its 16S sequence known?
1.5 Summary
After successfully working through this section you should be able to:
- use
BLASTandsilvato obtain taxonomic identity from 16S sequence data - interpret
BLASTandSILVAoutput - explain the difference in results between
BLASTandsilvataxonomy assignment

