13 Did SARS-CoV-2 Change During the COVID-19 Pandemic?
As the pandemic raged around the world (Figure 13.1), increasing numbers of people became infected.
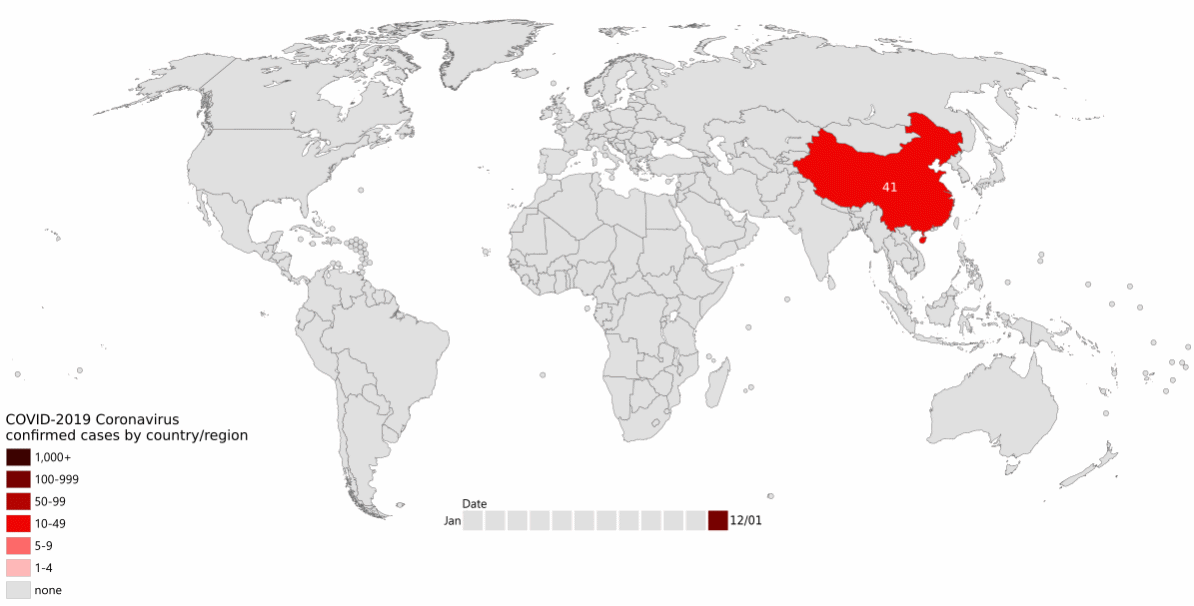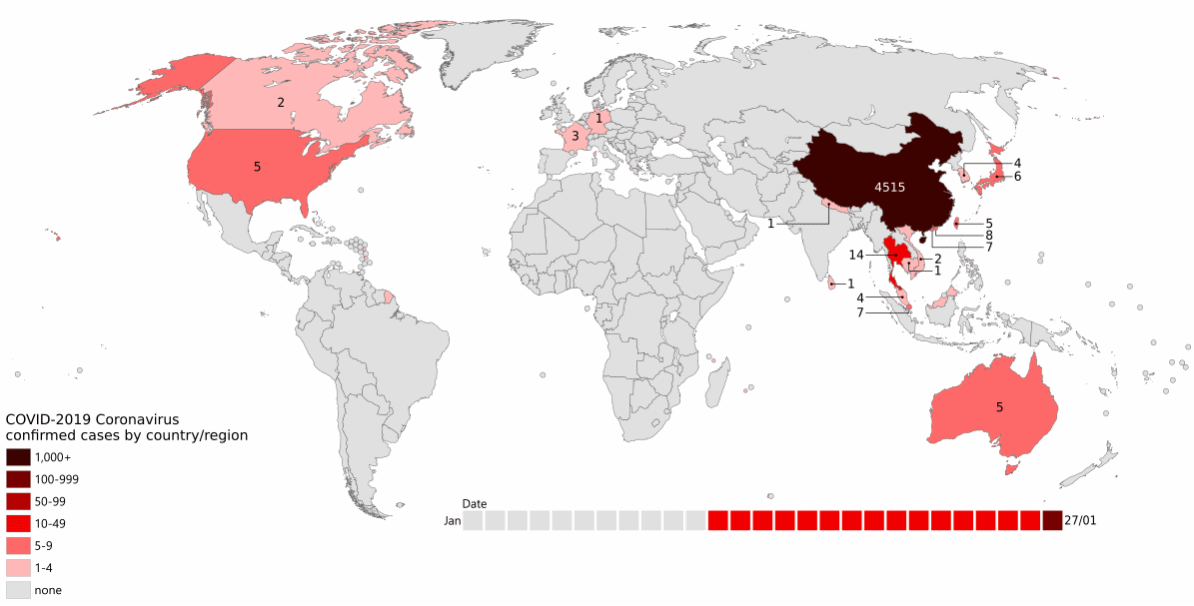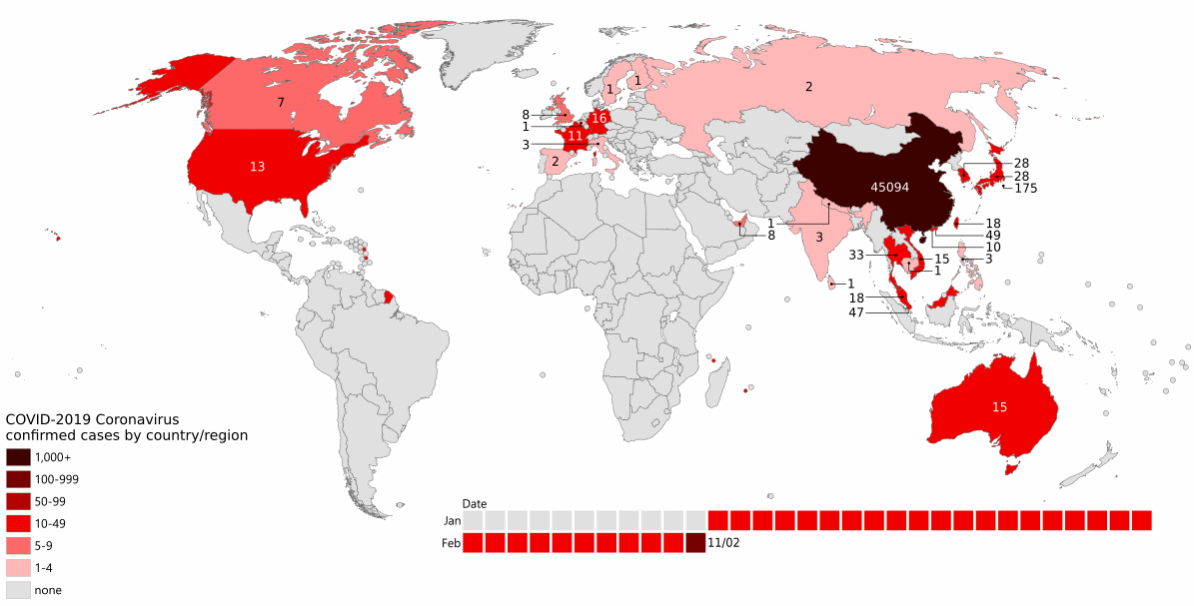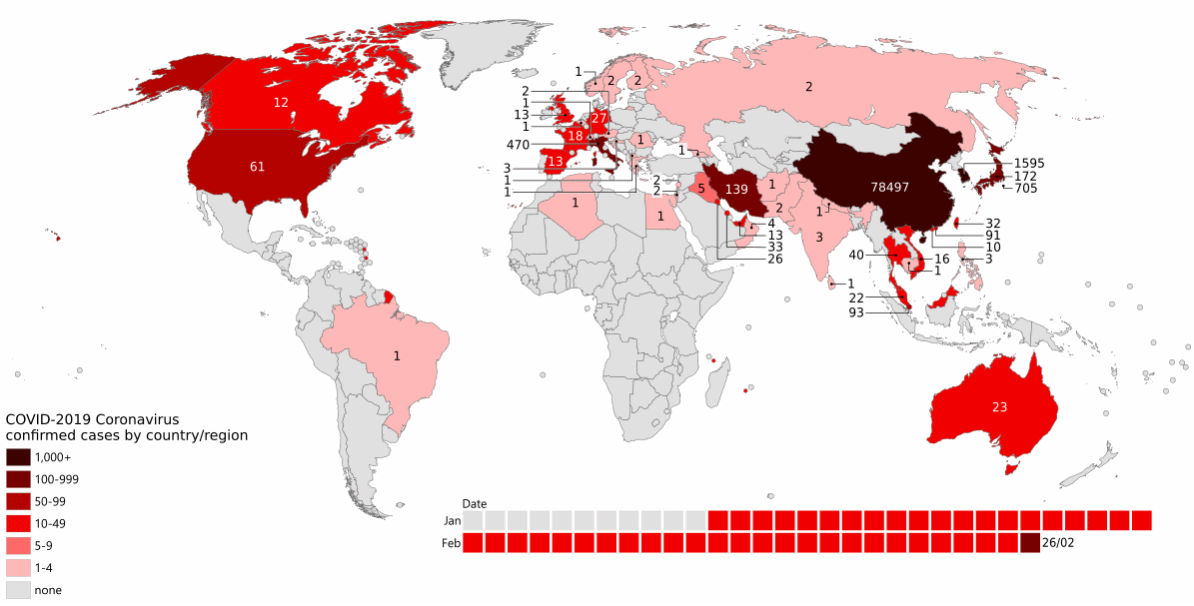
More infections meant more virus, which implies more viral replication and individual virus particles. This in turn implies more opportunities for mistakes during reproduction: more opportunities for mutation to occur. There was both a larger population of virus, and a larger effective population of the virus.
Any of the resulting sequence variants might have an important effect, maybe changing the clinical profile of the virus - its virulence, its residence time, ease of shedding, or some other characteristic. The variants might also provide a way to track transmission geographically, and over time.
We need you to identify and interpret SARS-CoV-2 sequence variants, and identify and interpret any changes that have occurred!
13.1 Your challenge
We need you to:
- Use the Oxford Nanopore (ONT) reads from the Sutch SARS-CoV-2 isolate
ERR4164769 - Carry out quality control on the reads
- Call sequence variants between the Dutch isolate and your Wuhan 1 assembly
- Report on the SNPs you find
- Map the ONT reads to your Wuhan 1 assembly and visualise them
- Be sure to visualise read quality with
FastQCand note any differences from the Wuhan 1 isolate reads - Remember - ONT reads are single-ended
- When considering SNPs
- How many are there?
- Do any lie within feature boundaries?
- What are the biological implications of changes to that feature’s product?
- You will need to increase the
maximum size of bam chunkssetting inJBrowseto visualise ONT reads - How do the ONT reads look different, visually, from Illumina data?
Please complete the formative quiz on MyPlace to earn your bioinformatics wings!